Systems Biology Markup Language (SBML)

Systems Biology Markup Language (SBML)
The Systems Biology Markup Language (SBML) is standard format for expressing dynamic models in computational biology. APM is a modeling language that enables the use of large-scale SBML models for dynamic simulation, parameter estimation, and optimization.
A large collection of SBML models is found in the Biomodels Database. To use these models in APM, they must first be converted with the following utility. Once converted, the model can be simulated through APM MATLAB, APM Python, or through a Web Interface.
SBFC Converter to APM (Beta Version)
Source code for the SBML2APM converter is available as part of SBFC: System Biology Format Converter, which is an open-source project aimed to provide a generic framework that potentially allows conversion between any two formats. The SBFC utilizes JSBML, a free and open-source java programming library to read, write, manipulate, translate, and validate SBML files and data streams. JSBML is a library that can be embedded in custom applications.
Example Applications from the BioModels Database
- Komarova2003_BoneRemodeling - BIOMD0000000148 - SBML XML Model
- Fisher2006_NFAT_Activation - BIOMD0000000123 - SBML XML Model
- Restif2007_Vaccination_Invasion - BIOMD0000000294 - SBML XML Model
Fig 1. Select sequential simulation and click the green run button. Solution results appear below.
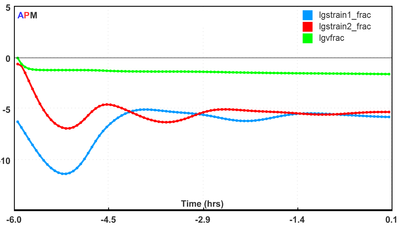
Fig 2. Trends of the model response are available through the web-interface.
References
- Lewis, N.R., Hedengren, J.D., Haseltine, E.L., Hybrid Dynamic Optimization Methods for Systems Biology with Efficient Sensitivities, Special Issue on Algorithms and Applications in Dynamic Optimization, Processes, 2015, 3(3), 701-729; doi:10.3390/pr3030701. Article | Source Code
- Safdarnejad, S.M., Hedengren, J.D., Lewis, N.R., Haseltine, E., Initialization Strategies for Optimization of Dynamic Systems, Computers and Chemical Engineering, 2015, Vol. 78, pp. 39-50, DOI: 10.1016/j.compchemeng.2015.04.016. Article
